Perform a principal components analysis (PCA) on a matrix or data frame and
return a pcomp object.
pcomp(x, ...)
# S3 method for class 'formula'
pcomp(formula, data = NULL, subset, na.action, method = c("svd", "eigen"), ...)
# Default S3 method
pcomp(
x,
method = c("svd", "eigen"),
scores = TRUE,
center = TRUE,
scale = TRUE,
tol = NULL,
covmat = NULL,
subset = rep(TRUE, nrow(as.matrix(x))),
...
)
# S3 method for class 'pcomp'
print(x, ...)
# S3 method for class 'pcomp'
summary(object, loadings = TRUE, cutoff = 0.1, ...)
# S3 method for class 'summary.pcomp'
print(x, digits = 3, loadings = x$print.loadings, cutoff = x$cutoff, ...)
# S3 method for class 'pcomp'
plot(
x,
which = c("screeplot", "loadings", "correlations", "scores"),
choices = 1L:2L,
col = par("col"),
bar.col = "gray",
circle.col = "gray",
ar.length = 0.1,
pos = NULL,
labels = NULL,
cex = par("cex"),
main = paste(deparse(substitute(x)), which, sep = " - "),
xlab,
ylab,
...
)
# S3 method for class 'pcomp'
screeplot(
x,
npcs = min(10, length(x$sdev)),
type = c("barplot", "lines"),
col = "cornsilk",
main = deparse(substitute(x)),
...
)
# S3 method for class 'pcomp'
points(
x,
choices = 1L:2L,
type = "p",
pch = par("pch"),
col = par("col"),
bg = par("bg"),
cex = par("cex"),
...
)
# S3 method for class 'pcomp'
lines(
x,
choices = 1L:2L,
groups,
type = c("p", "e"),
col = par("col"),
border = par("fg"),
level = 0.9,
...
)
# S3 method for class 'pcomp'
text(
x,
choices = 1L:2L,
labels = NULL,
col = par("col"),
cex = par("cex"),
pos = NULL,
...
)
# S3 method for class 'pcomp'
biplot(x, choices = 1L:2L, scale = 1, pc.biplot = FALSE, ...)
# S3 method for class 'pcomp'
pairs(
x,
choices = 1L:3L,
type = c("loadings", "correlations"),
col = par("col"),
circle.col = "gray",
ar.col = par("col"),
ar.length = 0.05,
pos = NULL,
ar.cex = par("cex"),
cex = par("cex"),
...
)
# S3 method for class 'pcomp'
predict(object, newdata, dim = length(object$sdev), ...)
# S3 method for class 'pcomp'
correlation(x, newvars, dim = length(x$sdev), ...)
scores(x, ...)
# S3 method for class 'pcomp'
scores(x, labels = NULL, dim = length(x$sdev), ...)Arguments
- x
A matrix or data frame with numeric data.
- ...
Arguments passed to or from other methods. If
xis a formula one might specifyscale,tolorcovmat.- formula
A formula with no response variable, referring only to numeric variables.
- data
An optional data frame (or similar, see
stats::model.frame()) containing the variables in the formula. By default the variables are taken fromenvironment(formula).- subset
An optional vector used to select rows (observations) of the data matrix
x.- na.action
A function which indicates what should happen when the data contain
NAs. The default is set by thena.actionsetting ofoptions(), and isstats::na.fail()if that is not set. The 'factory-fresh' default isstats::na.omit().- method
Either
"svd"(usingstats::prcomp()),"eigen"(usingstats::princomp()), or an abbreviation.- scores
A logical value indicating whether the score on each principal component should be calculated.
- center
A logical value indicating whether the variables should centered. Alternately, a vector of length equal the number of columns of
xcan be supplied. The value is passed toscale. Note that this argument is ignored formethod = "eigen"and the dataset is always centered in this case.- scale
A logical value indicating whether the variables should be scaled to have unit variance before the analysis takes place. The default is
TRUE, which in general, is advisable. Alternatively, a vector of length equal the number of columns ofxcan be supplied. The value is passed toscale().- tol
Only when
method = "svd". A value indicating the magnitude below which components should be omitted. (Components are omitted if their standard deviations are less than or equal totoltimes the standard deviation of the first component.) With the default null setting, no components are omitted. Other settings fortol =could betol = 0ortol = sqrt(.Machine$double.eps), which would omit essentially constant components.- covmat
A covariance matrix, or a covariance list as returned by
stats::cov.wt()(andMASS::cov.mve()orMASS::cov.mcd()from package MASS). If supplied, this is used rather than the covariance matrix ofx.- object
A 'pcomp' object.
- loadings
Do we also summarize the loadings?
- cutoff
The cutoff value below which loadings are replaced by white spaces in the table. That way, larger values are easier to spot and to read in large tables.
- digits
The number of digits to print.
- which
The graph to plot.
- choices
Which principal axes to plot. For 2D graphs, specify two integers.
- col
The color to use in graphs.
- bar.col
The color of bars in the screeplot.
- circle.col
The color for the circle in the loadings or correlations plots.
- ar.length
The length of the arrows in the loadings and correlations plots.
- pos
The position of text relative to arrows in loadings and correlation plots.
- labels
The labels to write. If
NULLdefault values are computed.- cex
The factor of expansion for text (labels) in the graphs.
- main
The title of the graph.
- xlab
The label of the x-axis.
- ylab
The label of the y-axis.
- npcs
The number of principal components to represent in the screeplot.
- type
The type of screeplot (
"barplot"or"lines") or pairs plot ("loadings"or"correlations").- pch
The type of symbol to use.
- bg
The background color for symbols.
- groups
A grouping factor.
- border
The color of the border.
- level
The probability level to use to draw the ellipse.
- pc.biplot
Do we create a Gabriel's biplot (see
stats::biplot())?- ar.col
Color of arrows.
- ar.cex
Expansion factor for text on arrows.
- newdata
New individuals with observations for the same variables as those used for calculating the PCA. You can then plot these additional individuals in the scores plot.
- dim
The number of principal components to keep.
- newvars
New variables with observations for same individuals as those used for calculating the PCA. Correlation with PCs is calculated. You can then plot these additional variables in the correlation plot.
Value
A c("pcomp", "pca", "princomp") object.
Details
pcomp() is a generic function with "formula" and "default"
methods. It is essentially a wrapper around stats::prcomp() and
stats::princomp() to provide a coherent interface and object for both
methods.
A 'pcomp' object is created. It inherits from 'pca' (as in labdsv package, but not compatible with the version of 'pca' in ade4) and of 'princomp'.
For more information on algorithms, refer to stats::prcomp() for
method = "svd" or stats::princomp() for method = "eigen".
Note
The signs of the columns for the loadings and scores are arbitrary. So, they could differ between functions for PCA, and even between different builds of R.
Examples
# Let's analyze mtcars without the Mercedes data (rows 8:14)
data(mtcars)
cars.pca <- pcomp(~ mpg + cyl + disp + hp + drat + wt + qsec,
data = mtcars, subset = -(8:14))
cars.pca
#> Call:
#> pcomp(formula = ~mpg + cyl + disp + hp + drat + wt + qsec, data = mtcars,
#> subset = -(8:14))
#>
#> Variances:
#> PC1 PC2 PC3 PC4 PC5 PC6 PC7
#> 5.13759552 1.21698212 0.28325478 0.15620899 0.12409321 0.05604916 0.02581622
#>
#> 7 variables and 25 observations.
summary(cars.pca)
#> Importance of components (eigenvalues):
#> PC1 PC2 PC3 PC4 PC5 PC6 PC7
#> Variance 5.138 1.217 0.2833 0.1562 0.1241 0.05605 0.02582
#> Proportion of Variance 0.734 0.174 0.0405 0.0223 0.0177 0.00801 0.00369
#> Cumulative Proportion 0.734 0.908 0.9483 0.9706 0.9883 0.99631 1.00000
#>
#> Loadings (eigenvectors, rotation matrix):
#> PC1 PC2 PC3 PC4 PC5 PC6 PC7
#> mpg -0.415 0.107 -0.754 -0.353 -0.318 0.144
#> cyl 0.425 0.165 -0.447 0.289 0.485 0.521
#> disp 0.423 0.110 -0.234 -0.465 0.103 -0.726
#> hp 0.385 -0.349 -0.106 -0.817 0.203
#> drat -0.320 -0.505 -0.736 0.208 0.222
#> wt 0.400 0.262 -0.499 -0.590 0.416
#> qsec -0.240 0.733 -0.323 -0.267 0.475
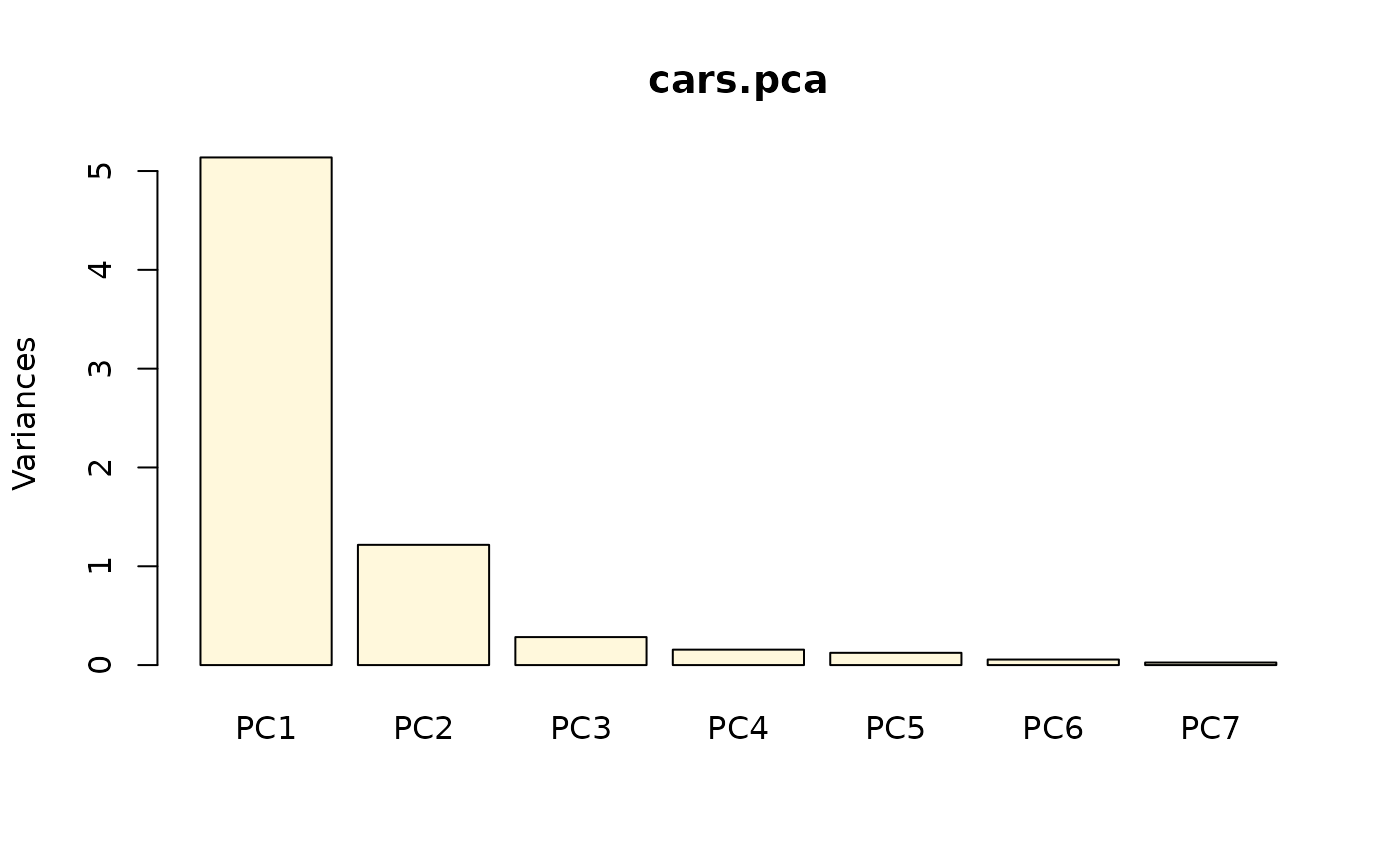
screeplot(cars.pca)
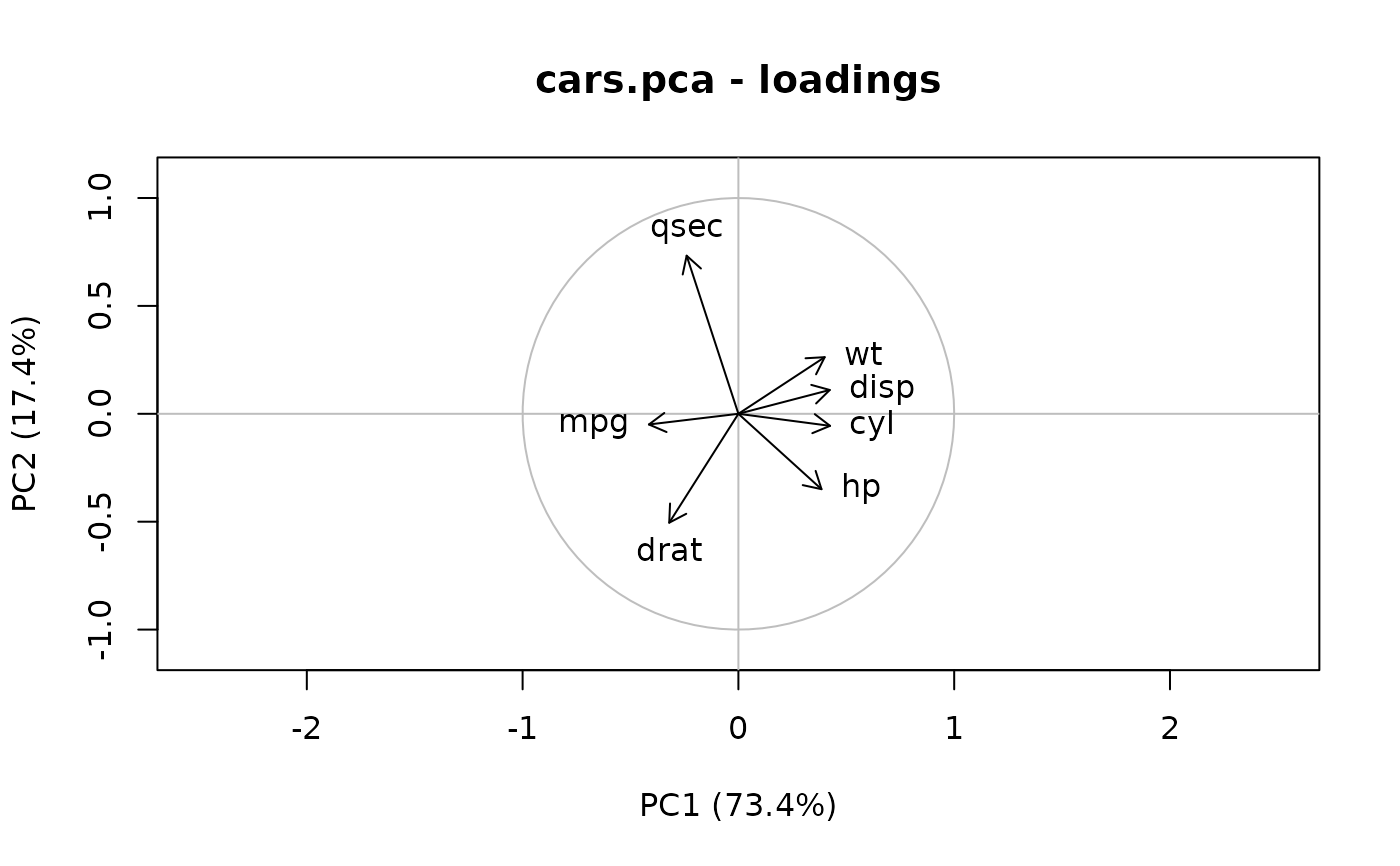
 # Loadings are extracted and plotted this way:
(cars.ldg <- loadings(cars.pca))
#>
#> Loadings:
#> PC1 PC2 PC3 PC4 PC5 PC6 PC7
#> mpg -0.415 0.107 -0.754 -0.353 -0.318 0.144
#> cyl 0.425 0.165 -0.447 0.289 0.485 0.521
#> disp 0.423 0.110 -0.234 -0.465 0.103 -0.726
#> hp 0.385 -0.349 -0.106 -0.817 0.203
#> drat -0.320 -0.505 -0.736 0.208 0.222
#> wt 0.400 0.262 -0.499 -0.590 0.416
#> qsec -0.240 0.733 -0.323 -0.267 0.475
#>
#> PC1 PC2 PC3 PC4 PC5 PC6 PC7
#> SS loadings 1.000 1.000 1.000 1.000 1.000 1.000 1.000
#> Proportion Var 0.143 0.143 0.143 0.143 0.143 0.143 0.143
#> Cumulative Var 0.143 0.286 0.429 0.571 0.714 0.857 1.000
plot(cars.pca, which = "loadings") # Equivalent to vectorplot(cars.ldg)
# Loadings are extracted and plotted this way:
(cars.ldg <- loadings(cars.pca))
#>
#> Loadings:
#> PC1 PC2 PC3 PC4 PC5 PC6 PC7
#> mpg -0.415 0.107 -0.754 -0.353 -0.318 0.144
#> cyl 0.425 0.165 -0.447 0.289 0.485 0.521
#> disp 0.423 0.110 -0.234 -0.465 0.103 -0.726
#> hp 0.385 -0.349 -0.106 -0.817 0.203
#> drat -0.320 -0.505 -0.736 0.208 0.222
#> wt 0.400 0.262 -0.499 -0.590 0.416
#> qsec -0.240 0.733 -0.323 -0.267 0.475
#>
#> PC1 PC2 PC3 PC4 PC5 PC6 PC7
#> SS loadings 1.000 1.000 1.000 1.000 1.000 1.000 1.000
#> Proportion Var 0.143 0.143 0.143 0.143 0.143 0.143 0.143
#> Cumulative Var 0.143 0.286 0.429 0.571 0.714 0.857 1.000
plot(cars.pca, which = "loadings") # Equivalent to vectorplot(cars.ldg)
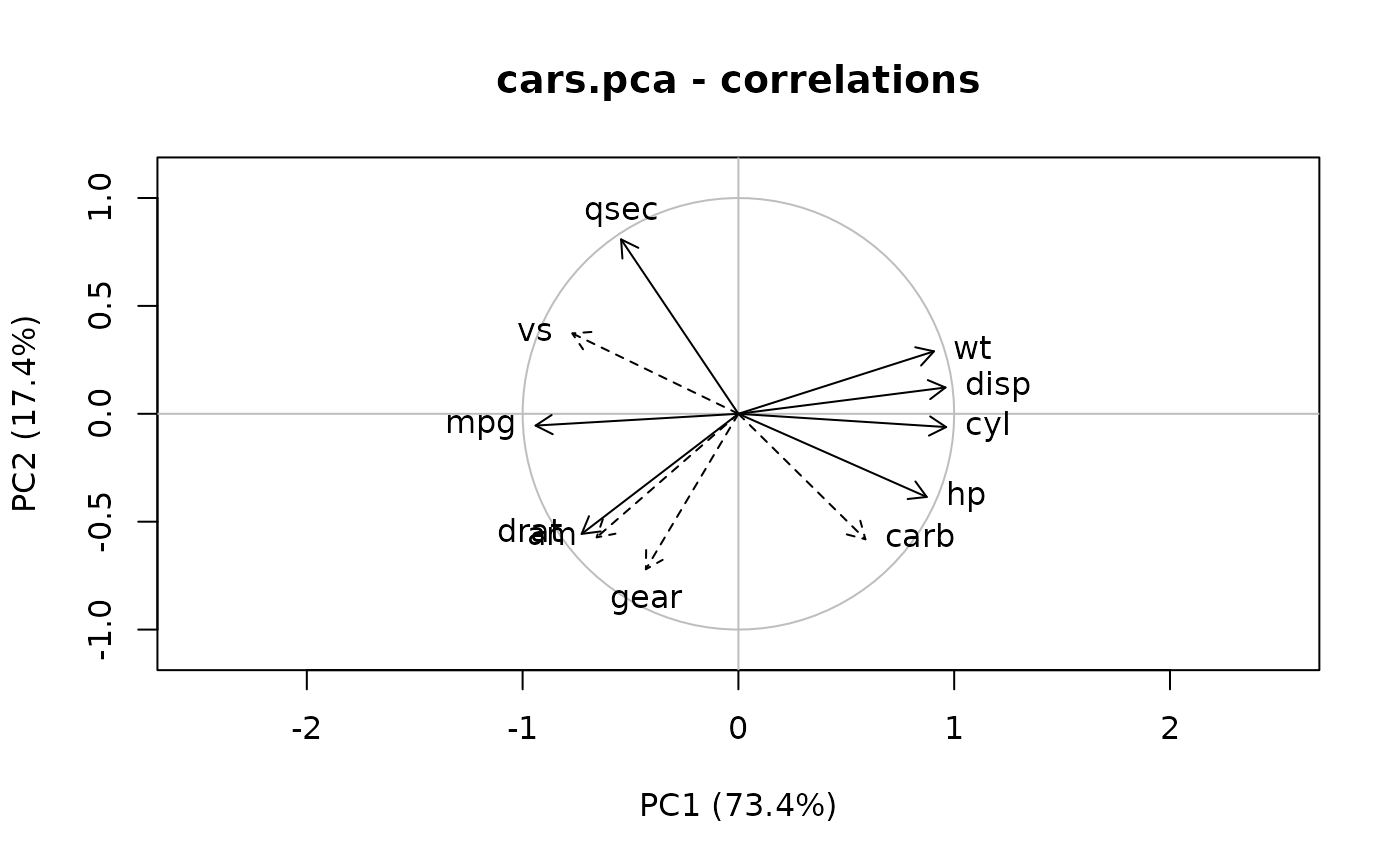
 # Similarly, correlations of variables with PCs are extracted and plotted:
(cars.cor <- Correlation(cars.pca))
#> Matrix of PCA variables and components correlation:
#> PC1 PC2 PC3 PC4 PC5 PC6 PC7
#> mpg -0.940 -0.055 0.057 -0.298 -0.124 -0.075 0.023
#> cyl 0.963 -0.062 0.088 -0.177 0.102 0.115 0.084
#> disp 0.960 0.122 -0.124 -0.184 0.036 0.003 -0.117
#> hp 0.873 -0.385 -0.056 0.039 -0.288 0.048 0.005
#> drat -0.726 -0.557 -0.392 -0.030 0.073 0.053 0.009
#> wt 0.906 0.289 -0.266 0.006 0.004 -0.140 0.067
#> qsec -0.544 0.808 -0.172 -0.010 -0.094 0.112 0.010
plot(cars.pca, which = "correlations") # Equivalent to vectorplot(cars.cor)
# One can add supplementary variables on this graph
lines(Correlation(cars.pca,
newvars = mtcars[-(8:14), c("vs", "am", "gear", "carb")]))
# Similarly, correlations of variables with PCs are extracted and plotted:
(cars.cor <- Correlation(cars.pca))
#> Matrix of PCA variables and components correlation:
#> PC1 PC2 PC3 PC4 PC5 PC6 PC7
#> mpg -0.940 -0.055 0.057 -0.298 -0.124 -0.075 0.023
#> cyl 0.963 -0.062 0.088 -0.177 0.102 0.115 0.084
#> disp 0.960 0.122 -0.124 -0.184 0.036 0.003 -0.117
#> hp 0.873 -0.385 -0.056 0.039 -0.288 0.048 0.005
#> drat -0.726 -0.557 -0.392 -0.030 0.073 0.053 0.009
#> wt 0.906 0.289 -0.266 0.006 0.004 -0.140 0.067
#> qsec -0.544 0.808 -0.172 -0.010 -0.094 0.112 0.010
plot(cars.pca, which = "correlations") # Equivalent to vectorplot(cars.cor)
# One can add supplementary variables on this graph
lines(Correlation(cars.pca,
newvars = mtcars[-(8:14), c("vs", "am", "gear", "carb")]))
 # Plot the scores:
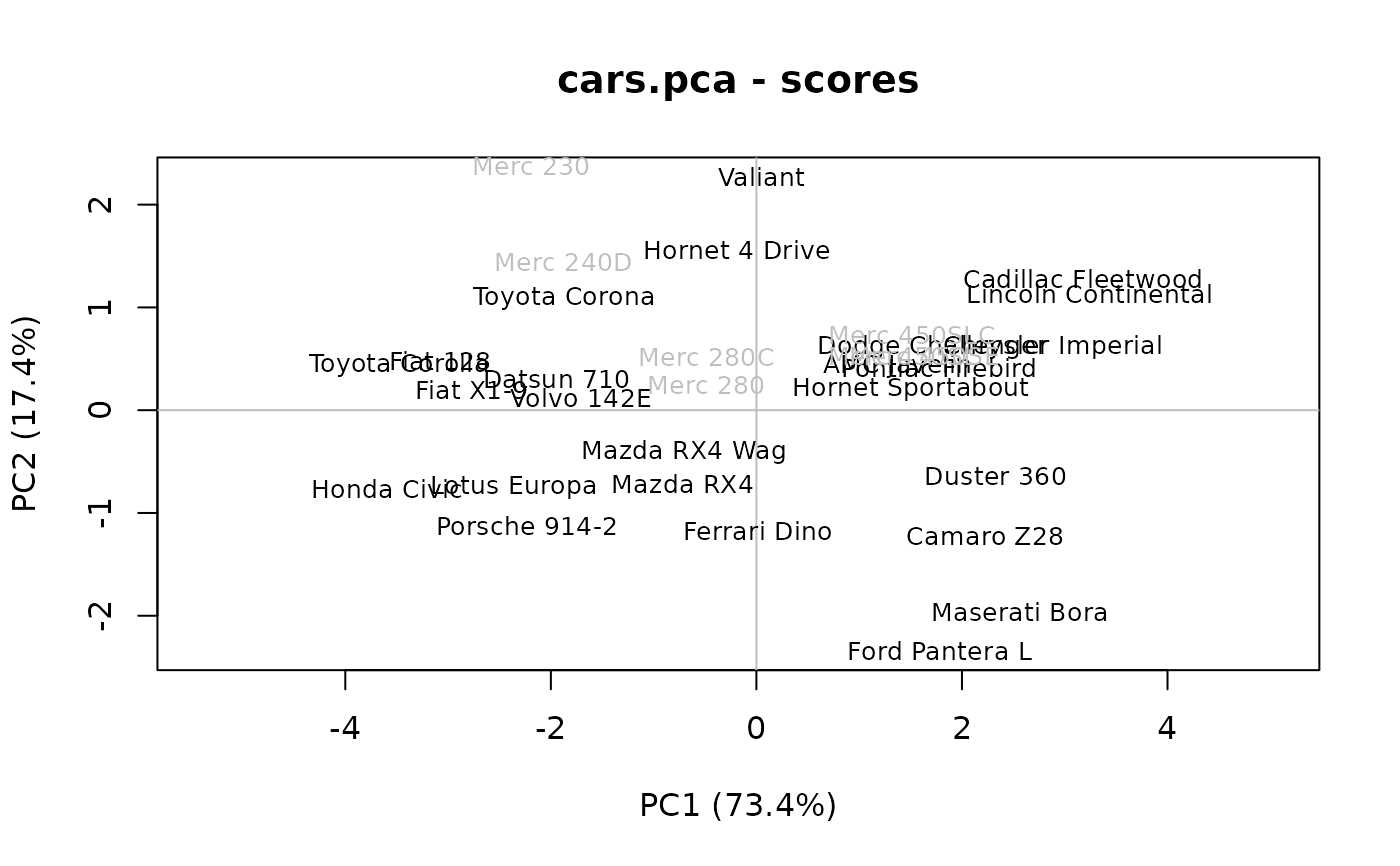
plot(cars.pca, which = "scores", cex = 0.8) # Similar to plot(scores(x)[, 1:2])
#> Warning: NAs introduced by coercion
# Add supplementary individuals to this plot (labels), also points() or lines()
text(predict(cars.pca, newdata = mtcars[8:14, ]),
labels = rownames(mtcars[8:14, ]), col = "gray", cex = 0.8)
# Plot the scores:
plot(cars.pca, which = "scores", cex = 0.8) # Similar to plot(scores(x)[, 1:2])
#> Warning: NAs introduced by coercion
# Add supplementary individuals to this plot (labels), also points() or lines()
text(predict(cars.pca, newdata = mtcars[8:14, ]),
labels = rownames(mtcars[8:14, ]), col = "gray", cex = 0.8)
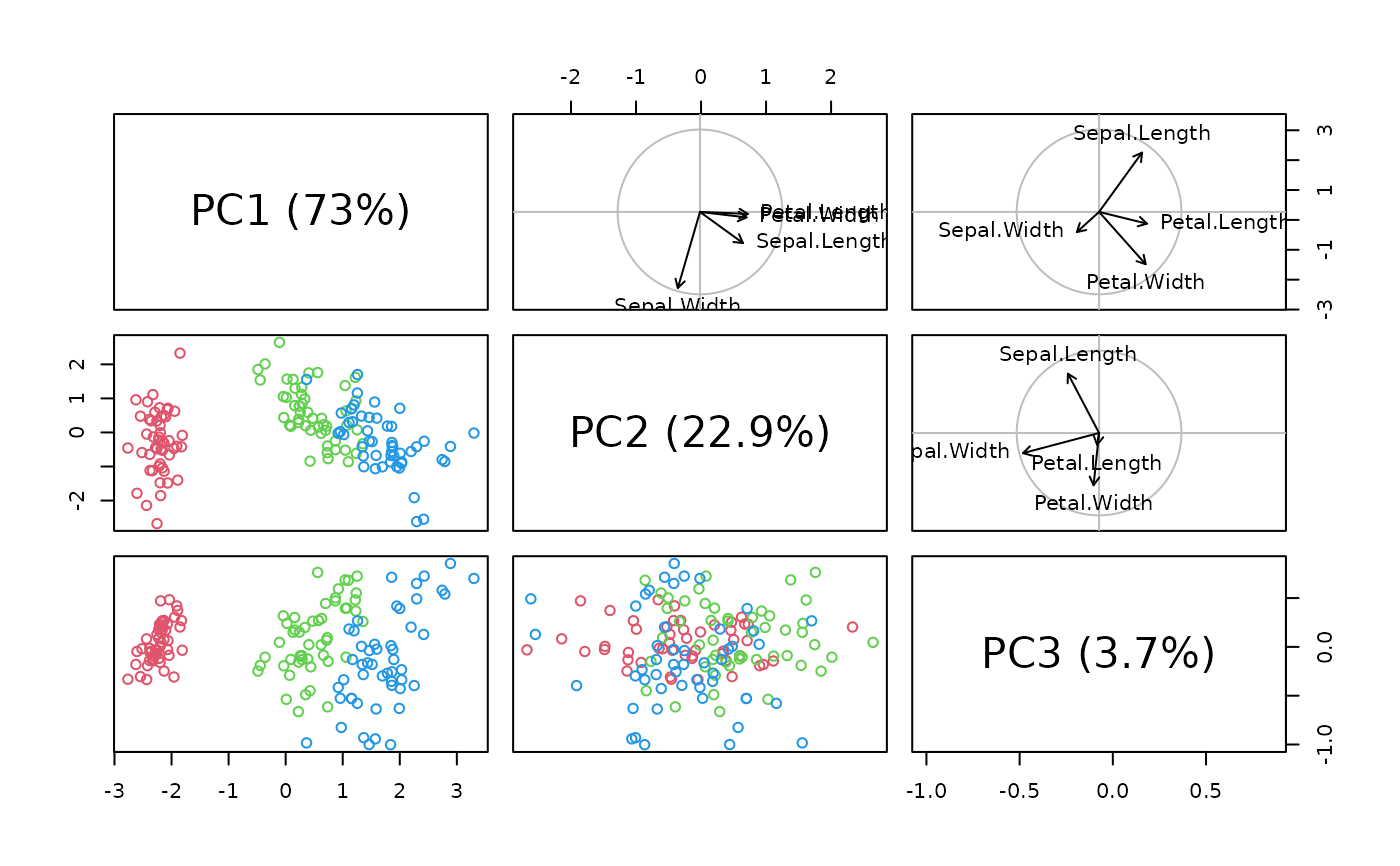
 # Pairs plot for 3 PCs
iris.pca <- pcomp(iris[, -5])
pairs(iris.pca, col = (2:4)[iris$Species])
# Pairs plot for 3 PCs
iris.pca <- pcomp(iris[, -5])
pairs(iris.pca, col = (2:4)[iris$Species])