
Time series decomposition using a regression model
decreg.RdProviding values coming from a regression on the original series, a tsd object is created using the original series, the regression model and the residuals
decreg(x, xreg, type="additive")Arguments
Value
a 'tsd' object
References
Frontier, S., 1981. Méthodes statistiques. Masson, Paris. 246 pp.
Kendall, M., 1976. Time-series. Charles Griffin & Co Ltd. 197 pp.
Legendre, L. & P. Legendre, 1984. Ecologie numérique. Tome 2: La structure des données écologiques. Masson, Paris. 335 pp.
Malinvaud, E., 1978. Méthodes statistiques de l'économétrie. Dunod, Paris. 846 pp.
Sokal, R.R. & F.J. Rohlf, 1981. Biometry. Freeman & Co, San Francisco. 860 pp.
Examples
data(marphy)
density <- ts(marphy[, "Density"])
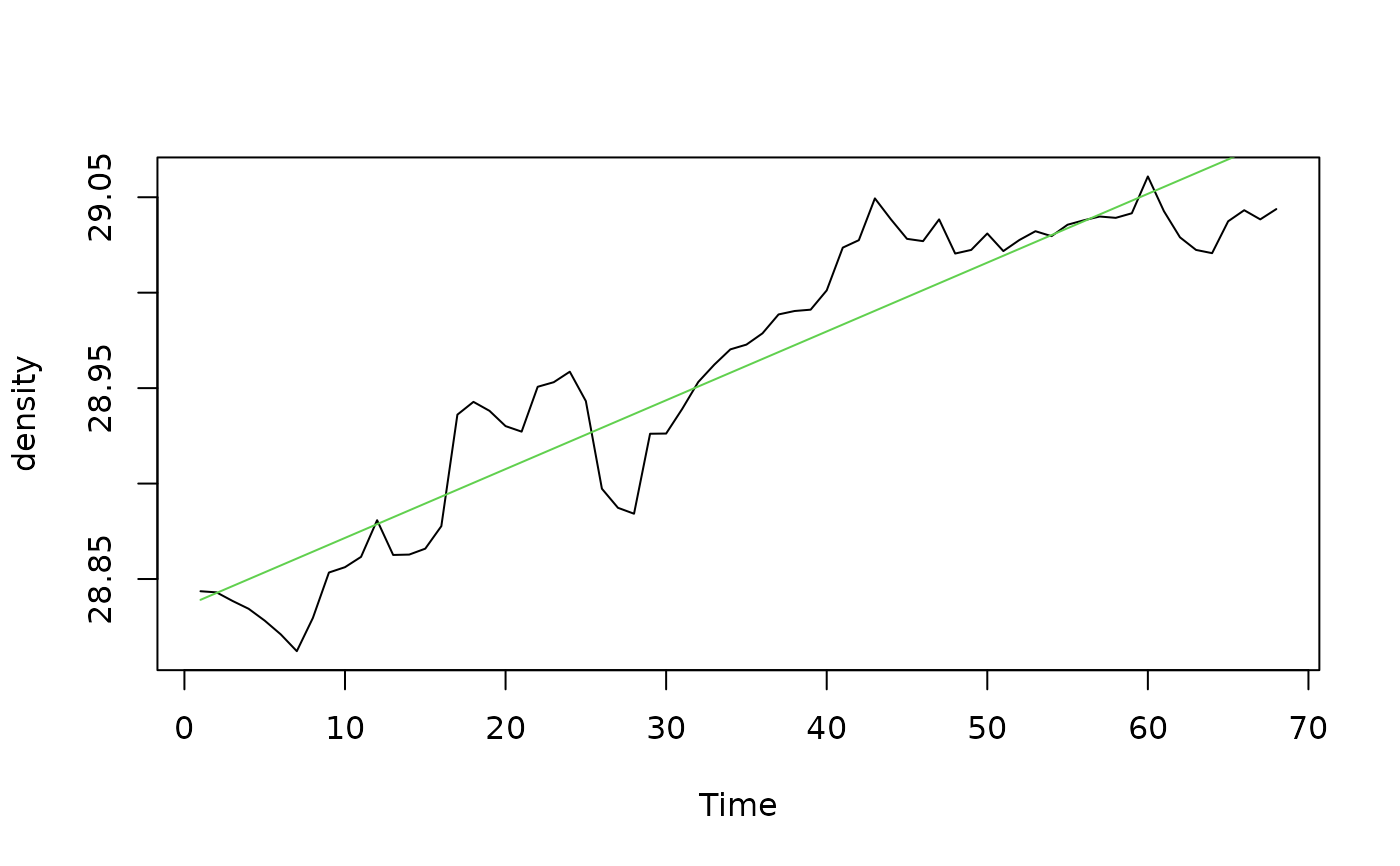
plot(density)
Time <- time(density)
# Linear model to represent trend
density.lin <- lm(density ~ Time)
summary(density.lin)
#>
#> Call:
#> lm(formula = density ~ Time)
#>
#> Residuals:
#> Min 1Q Median 3Q Max
#> -0.052226 -0.018014 0.001945 0.017672 0.058895
#>
#> Coefficients:
#> Estimate Std. Error t value Pr(>|t|)
#> (Intercept) 2.884e+01 6.585e-03 4379.27 <2e-16 ***
#> Time 3.605e-03 1.659e-04 21.73 <2e-16 ***
#> ---
#> Signif. codes: 0 ‘***’ 0.001 ‘**’ 0.01 ‘*’ 0.05 ‘.’ 0.1 ‘ ’ 1
#>
#> Residual standard error: 0.02685 on 66 degrees of freedom
#> Multiple R-squared: 0.8774, Adjusted R-squared: 0.8755
#> F-statistic: 472.3 on 1 and 66 DF, p-value: < 2.2e-16
#>
xreg <- predict(density.lin)
lines(xreg, col=3)
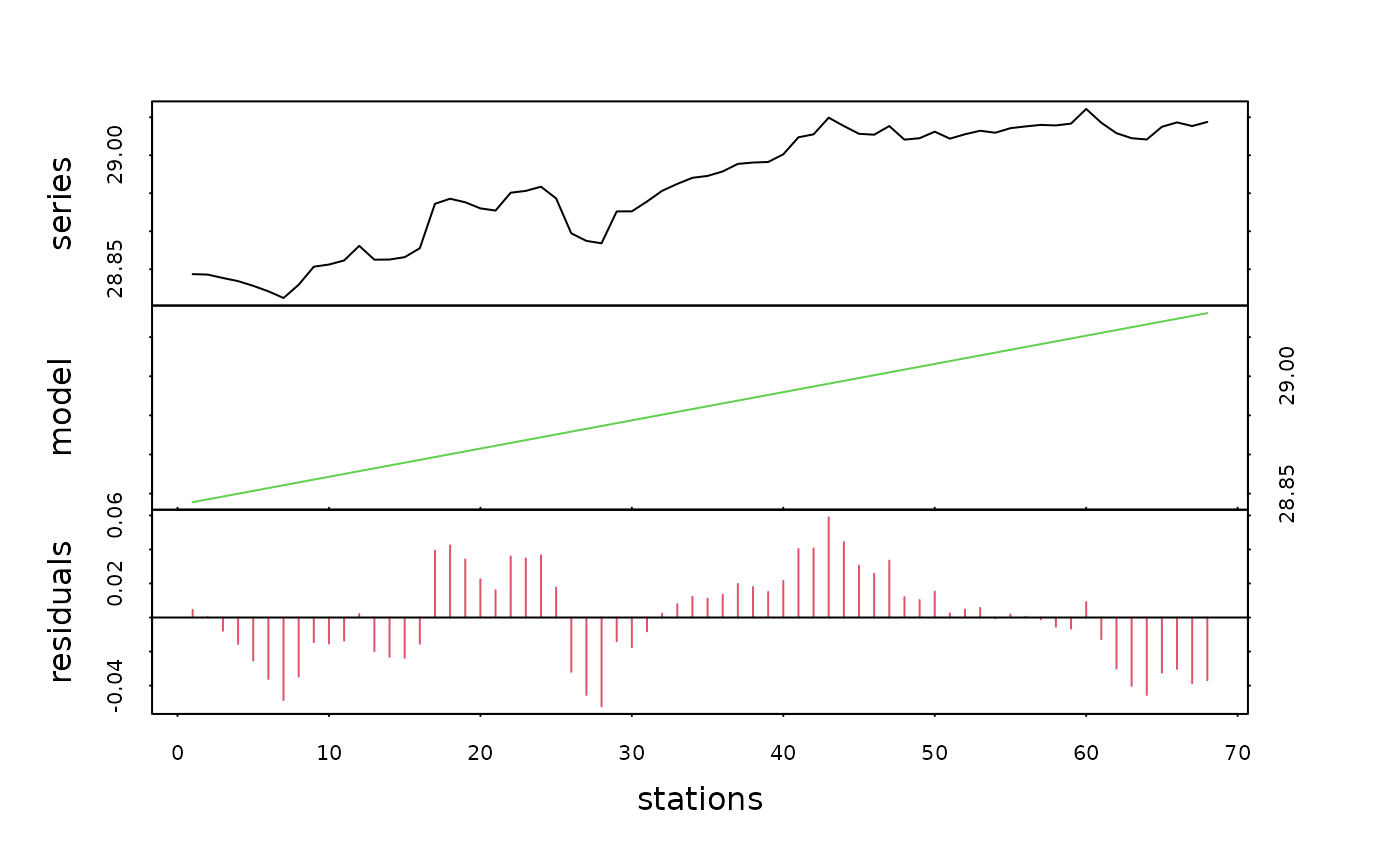
 density.dec <- decreg(density, xreg)
plot(density.dec, col=c(1, 3, 2), xlab="stations")
density.dec <- decreg(density, xreg)
plot(density.dec, col=c(1, 3, 2), xlab="stations")
 # Order 2 polynomial to represent trend
density.poly <- lm(density ~ Time + I(Time^2))
summary(density.poly)
#>
#> Call:
#> lm(formula = density ~ Time + I(Time^2))
#>
#> Residuals:
#> Min 1Q Median 3Q Max
#> -0.067079 -0.010665 0.001498 0.014777 0.045341
#>
#> Coefficients:
#> Estimate Std. Error t value Pr(>|t|)
#> (Intercept) 2.880e+01 8.374e-03 3439.404 < 2e-16 ***
#> Time 6.593e-03 5.600e-04 11.773 < 2e-16 ***
#> I(Time^2) -4.330e-05 7.866e-06 -5.505 6.73e-07 ***
#> ---
#> Signif. codes: 0 ‘***’ 0.001 ‘**’ 0.01 ‘*’ 0.05 ‘.’ 0.1 ‘ ’ 1
#>
#> Residual standard error: 0.02234 on 65 degrees of freedom
#> Multiple R-squared: 0.9164, Adjusted R-squared: 0.9138
#> F-statistic: 356.2 on 2 and 65 DF, p-value: < 2.2e-16
#>
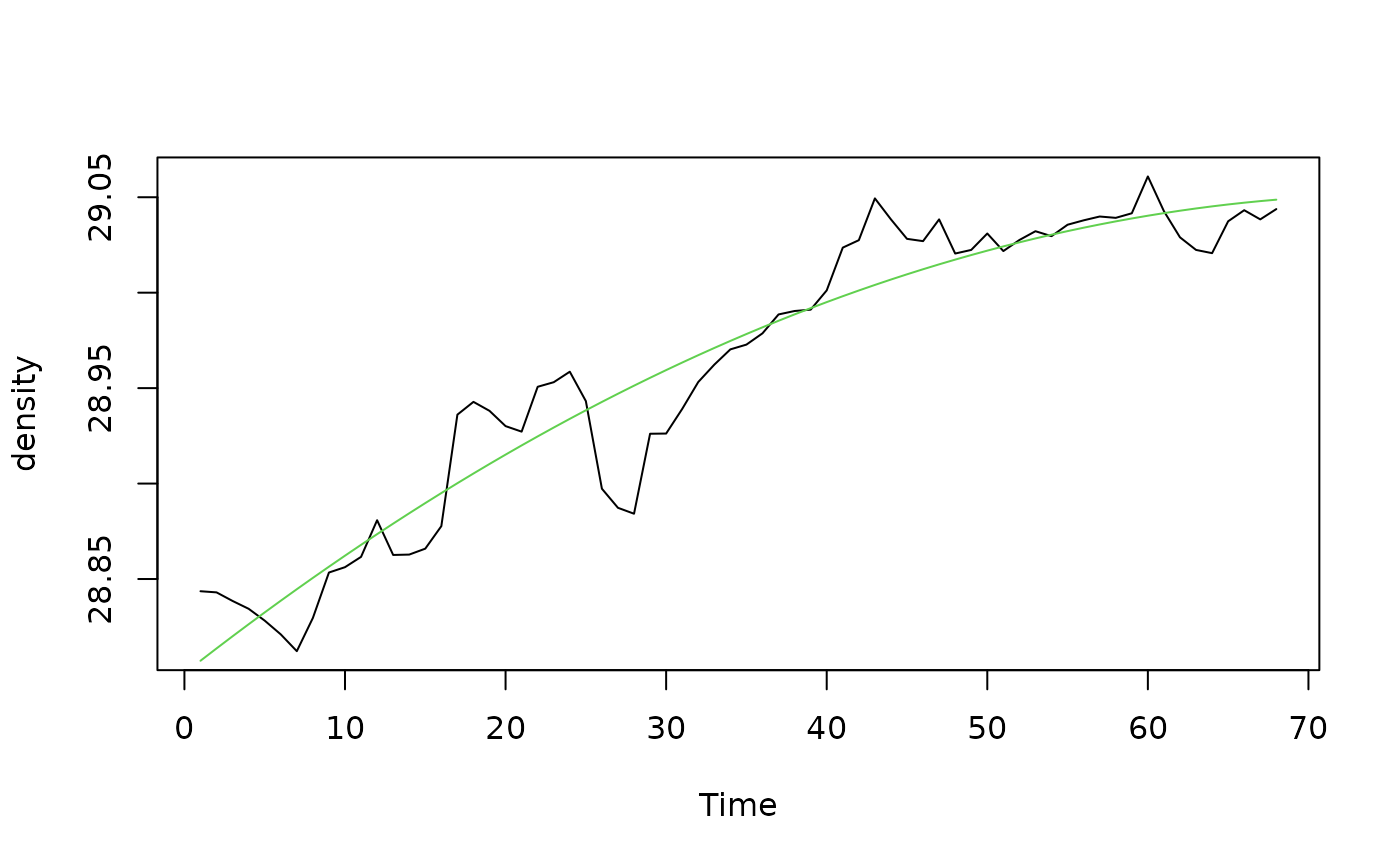
xreg2 <- predict(density.poly)
plot(density)
lines(xreg2, col=3)
# Order 2 polynomial to represent trend
density.poly <- lm(density ~ Time + I(Time^2))
summary(density.poly)
#>
#> Call:
#> lm(formula = density ~ Time + I(Time^2))
#>
#> Residuals:
#> Min 1Q Median 3Q Max
#> -0.067079 -0.010665 0.001498 0.014777 0.045341
#>
#> Coefficients:
#> Estimate Std. Error t value Pr(>|t|)
#> (Intercept) 2.880e+01 8.374e-03 3439.404 < 2e-16 ***
#> Time 6.593e-03 5.600e-04 11.773 < 2e-16 ***
#> I(Time^2) -4.330e-05 7.866e-06 -5.505 6.73e-07 ***
#> ---
#> Signif. codes: 0 ‘***’ 0.001 ‘**’ 0.01 ‘*’ 0.05 ‘.’ 0.1 ‘ ’ 1
#>
#> Residual standard error: 0.02234 on 65 degrees of freedom
#> Multiple R-squared: 0.9164, Adjusted R-squared: 0.9138
#> F-statistic: 356.2 on 2 and 65 DF, p-value: < 2.2e-16
#>
xreg2 <- predict(density.poly)
plot(density)
lines(xreg2, col=3)
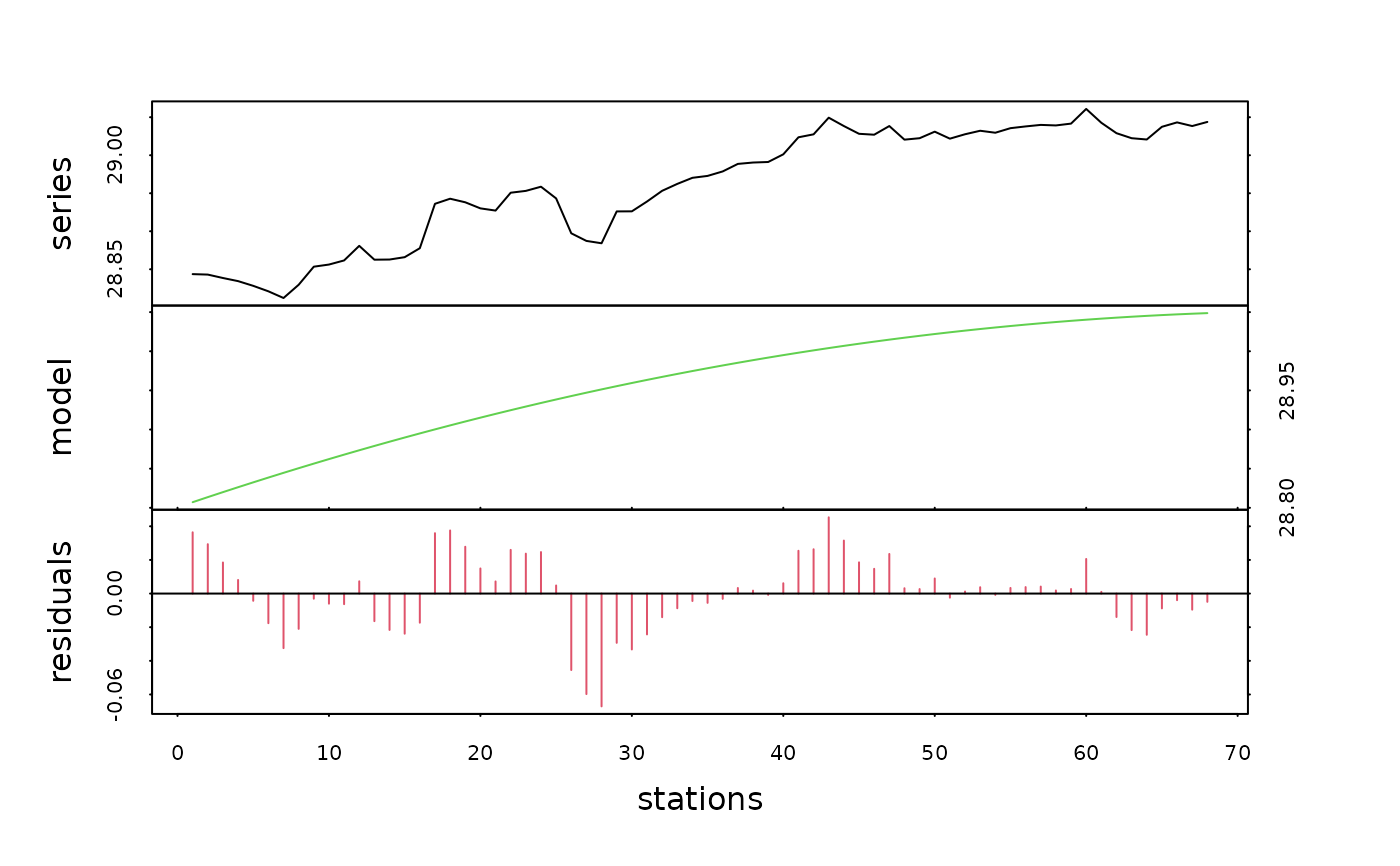
 density.dec2 <- decreg(density, xreg2)
plot(density.dec2, col=c(1, 3, 2), xlab="stations")
density.dec2 <- decreg(density, xreg2)
plot(density.dec2, col=c(1, 3, 2), xlab="stations")
 # Fit a sinusoidal model on seasonal (artificial) data
tser <- ts(sin((1:100)/12*pi)+rnorm(100, sd=0.3), start=c(1998, 4),
frequency=24)
Time <- time(tser)
tser.sin <- lm(tser ~ I(cos(2*pi*Time)) + I(sin(2*pi*Time)))
summary(tser.sin)
#>
#> Call:
#> lm(formula = tser ~ I(cos(2 * pi * Time)) + I(sin(2 * pi * Time)))
#>
#> Residuals:
#> Min 1Q Median 3Q Max
#> -0.75515 -0.14794 -0.01237 0.16597 0.82122
#>
#> Coefficients:
#> Estimate Std. Error t value Pr(>|t|)
#> (Intercept) 0.01511 0.03112 0.485 0.629
#> I(cos(2 * pi * Time)) -0.53156 0.04450 -11.944 <2e-16 ***
#> I(sin(2 * pi * Time)) 0.82928 0.04351 19.062 <2e-16 ***
#> ---
#> Signif. codes: 0 ‘***’ 0.001 ‘**’ 0.01 ‘*’ 0.05 ‘.’ 0.1 ‘ ’ 1
#>
#> Residual standard error: 0.3108 on 97 degrees of freedom
#> Multiple R-squared: 0.8364, Adjusted R-squared: 0.833
#> F-statistic: 248 on 2 and 97 DF, p-value: < 2.2e-16
#>
tser.reg <- predict(tser.sin)
tser.dec <- decreg(tser, tser.reg)
plot(tser.dec, col=c(1, 4), xlab="stations", stack=FALSE, resid=FALSE,
lpos=c(0, 4))
# Fit a sinusoidal model on seasonal (artificial) data
tser <- ts(sin((1:100)/12*pi)+rnorm(100, sd=0.3), start=c(1998, 4),
frequency=24)
Time <- time(tser)
tser.sin <- lm(tser ~ I(cos(2*pi*Time)) + I(sin(2*pi*Time)))
summary(tser.sin)
#>
#> Call:
#> lm(formula = tser ~ I(cos(2 * pi * Time)) + I(sin(2 * pi * Time)))
#>
#> Residuals:
#> Min 1Q Median 3Q Max
#> -0.75515 -0.14794 -0.01237 0.16597 0.82122
#>
#> Coefficients:
#> Estimate Std. Error t value Pr(>|t|)
#> (Intercept) 0.01511 0.03112 0.485 0.629
#> I(cos(2 * pi * Time)) -0.53156 0.04450 -11.944 <2e-16 ***
#> I(sin(2 * pi * Time)) 0.82928 0.04351 19.062 <2e-16 ***
#> ---
#> Signif. codes: 0 ‘***’ 0.001 ‘**’ 0.01 ‘*’ 0.05 ‘.’ 0.1 ‘ ’ 1
#>
#> Residual standard error: 0.3108 on 97 degrees of freedom
#> Multiple R-squared: 0.8364, Adjusted R-squared: 0.833
#> F-statistic: 248 on 2 and 97 DF, p-value: < 2.2e-16
#>
tser.reg <- predict(tser.sin)
tser.dec <- decreg(tser, tser.reg)
plot(tser.dec, col=c(1, 4), xlab="stations", stack=FALSE, resid=FALSE,
lpos=c(0, 4))
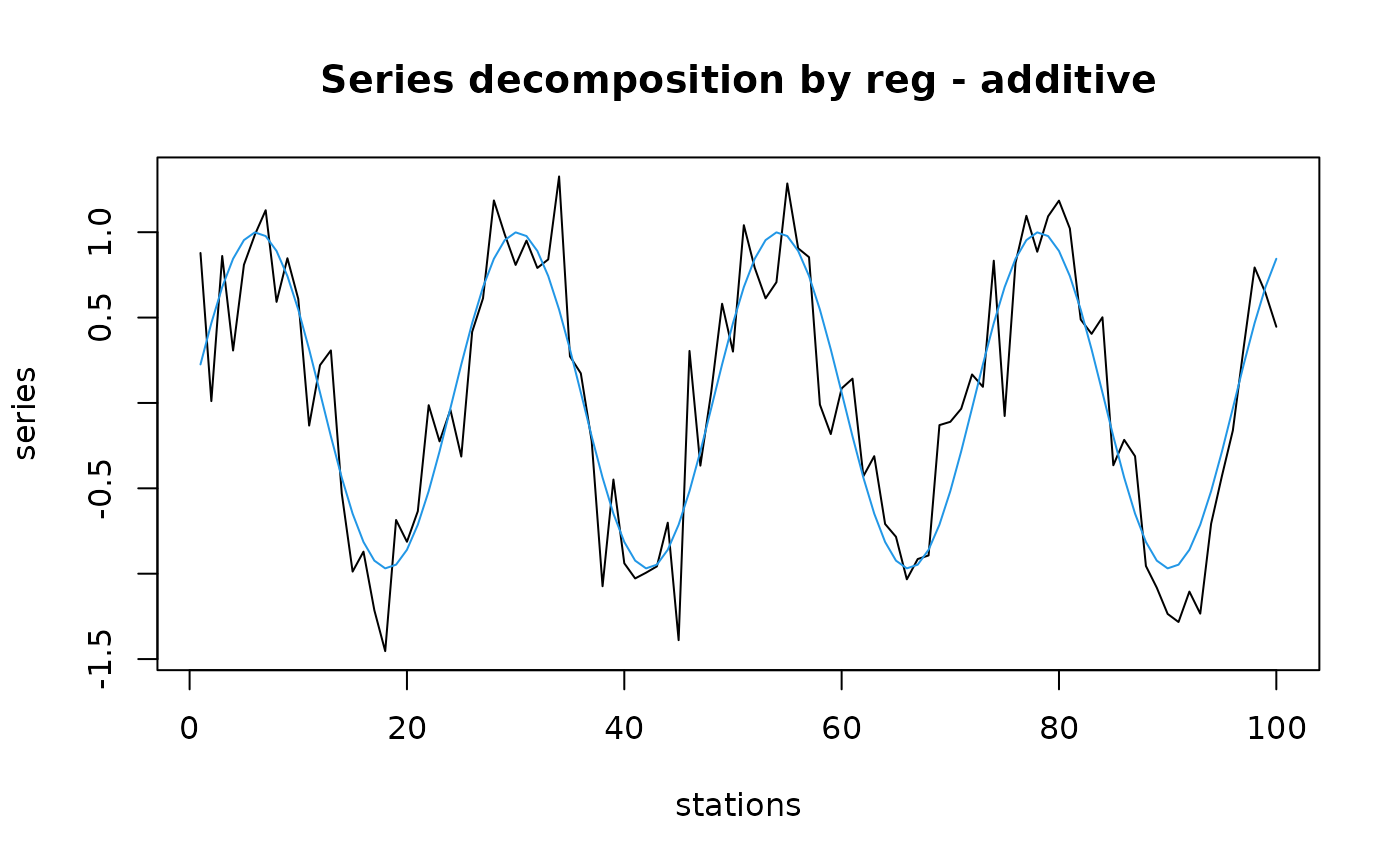 plot(tser.dec, col=c(1, 4, 2), xlab="stations")
plot(tser.dec, col=c(1, 4, 2), xlab="stations")
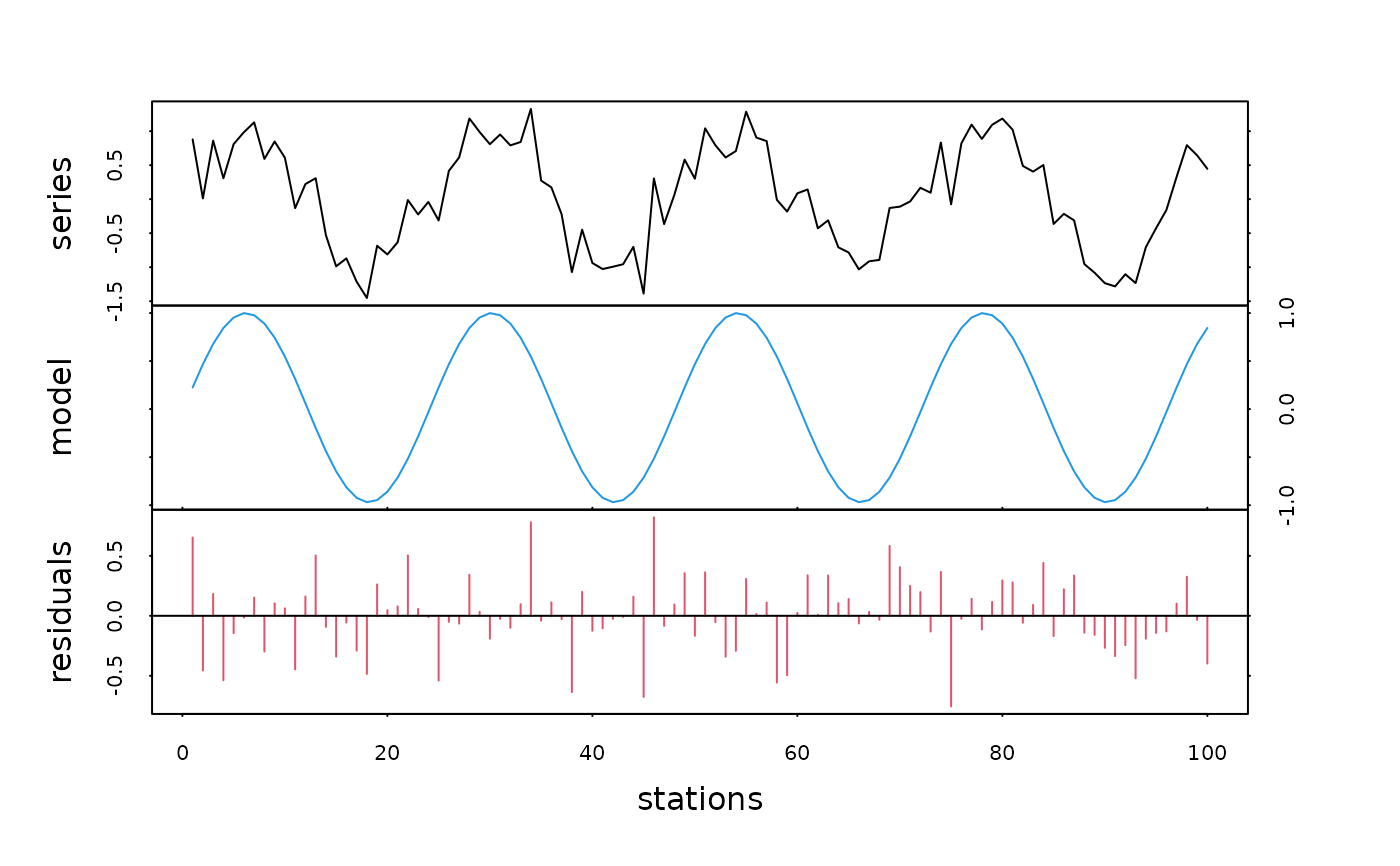 # One can also use nonlinear models (see 'nls')
# or autoregressive models (see 'ar' and others in 'ts' library)
# One can also use nonlinear models (see 'nls')
# or autoregressive models (see 'ar' and others in 'ts' library)
