
Supervised classification and regression using recursive partitioning
Source:R/ml_rpart.R
mlRpart.RdUnified (formula-based) interface version of the recursive partitioning
algorithm as implemented in rpart::rpart().
mlRpart(train, ...)
ml_rpart(train, ...)
# S3 method for class 'formula'
mlRpart(formula, data, ..., subset, na.action)
# Default S3 method
mlRpart(train, response, ..., .args. = NULL)
# S3 method for class 'mlRpart'
predict(
object,
newdata,
type = c("class", "membership", "both"),
method = c("direct", "cv"),
...
)Arguments
- train
a matrix or data frame with predictors.
- ...
further arguments passed to
rpart::rpart()or itspredict()method (see the corresponding help page.- formula
a formula with left term being the factor variable to predict (for supervised classification), a vector of numbers (for regression) and the right term with the list of independent, predictive variables, separated with a plus sign. If the data frame provided contains only the dependent and independent variables, one can use the
class ~ .short version (that one is strongly encouraged). Variables with minus sign are eliminated. Calculations on variables are possible according to usual formula convention (possibly protected by usingI()).- data
a data.frame to use as a training set.
- subset
index vector with the cases to define the training set in use (this argument must be named, if provided).
- na.action
function to specify the action to be taken if
NAs are found. Forml_rpart()na.failis used by default. The calculation is stopped if there is anyNAin the data. Another option isna.omit, where cases with missing values on any required variable are dropped (this argument must be named, if provided). For thepredict()method, the default, and most suitable option, isna.exclude. In that case, rows withNAs innewdata=are excluded from prediction, but reinjected in the final results so that the number of items is still the same (and in the same order asnewdata=).- response
a vector of factor (classification) or numeric (regression).
- .args.
used internally, do not provide anything here.
- object
an mlRpart object
- newdata
a new dataset with same conformation as the training set (same variables, except may by the class for classification or dependent variable for regression). Usually a test set, or a new dataset to be predicted.
- type
the type of prediction to return.
"class"by default, the predicted classes. Other options are"membership"the membership (number between 0 and 1) to the different classes, or"both"to return classes and memberships,- method
"direct"(default) or"cv"."direct"predicts new cases innewdata=if this argument is provided, or the cases in the training set if not. Take care that not providingnewdata=means that you just calculate the self-consistency of the classifier but cannot use the metrics derived from these results for the assessment of its performances. Either use a different data set innewdata=or use the alternate cross-validation ("cv") technique. If you specifymethod = "cv"thencvpredict()is used and you cannot providenewdata=in that case.
Value
ml_rpart()/mlRpart() creates an mlRpart, mlearning object
containing the classifier and a lot of additional metadata used by the
functions and methods you can apply to it like predict() or
cvpredict(). In case you want to program new functions or extract
specific components, inspect the "unclassed" object using unclass().
See also
mlearning(), cvpredict(), confusion(), also rpart::rpart()
that actually does the classification.
Examples
# Prepare data: split into training set (2/3) and test set (1/3)
data("iris", package = "datasets")
train <- c(1:34, 51:83, 101:133)
iris_train <- iris[train, ]
iris_test <- iris[-train, ]
# One case with missing data in train set, and another case in test set
iris_train[1, 1] <- NA
iris_test[25, 2] <- NA
iris_rpart <- ml_rpart(data = iris_train, Species ~ .)
summary(iris_rpart)
#> A mlearning object of class mlRpart (recursive partitioning tree):
#> Initial call: mlRpart.formula(formula = Species ~ ., data = iris_train)
#> n= 99
#>
#> node), split, n, loss, yval, (yprob)
#> * denotes terminal node
#>
#> 1) root 99 66 setosa (0.33333333 0.33333333 0.33333333)
#> 2) Petal.Length< 2.6 33 0 setosa (1.00000000 0.00000000 0.00000000) *
#> 3) Petal.Length>=2.6 66 33 versicolor (0.00000000 0.50000000 0.50000000)
#> 6) Petal.Width< 1.55 31 1 versicolor (0.00000000 0.96774194 0.03225806) *
#> 7) Petal.Width>=1.55 35 3 virginica (0.00000000 0.08571429 0.91428571) *
# Plot the decision tree for this classifier
plot(iris_rpart, margin = 0.03, uniform = TRUE)
text(iris_rpart, use.n = FALSE)
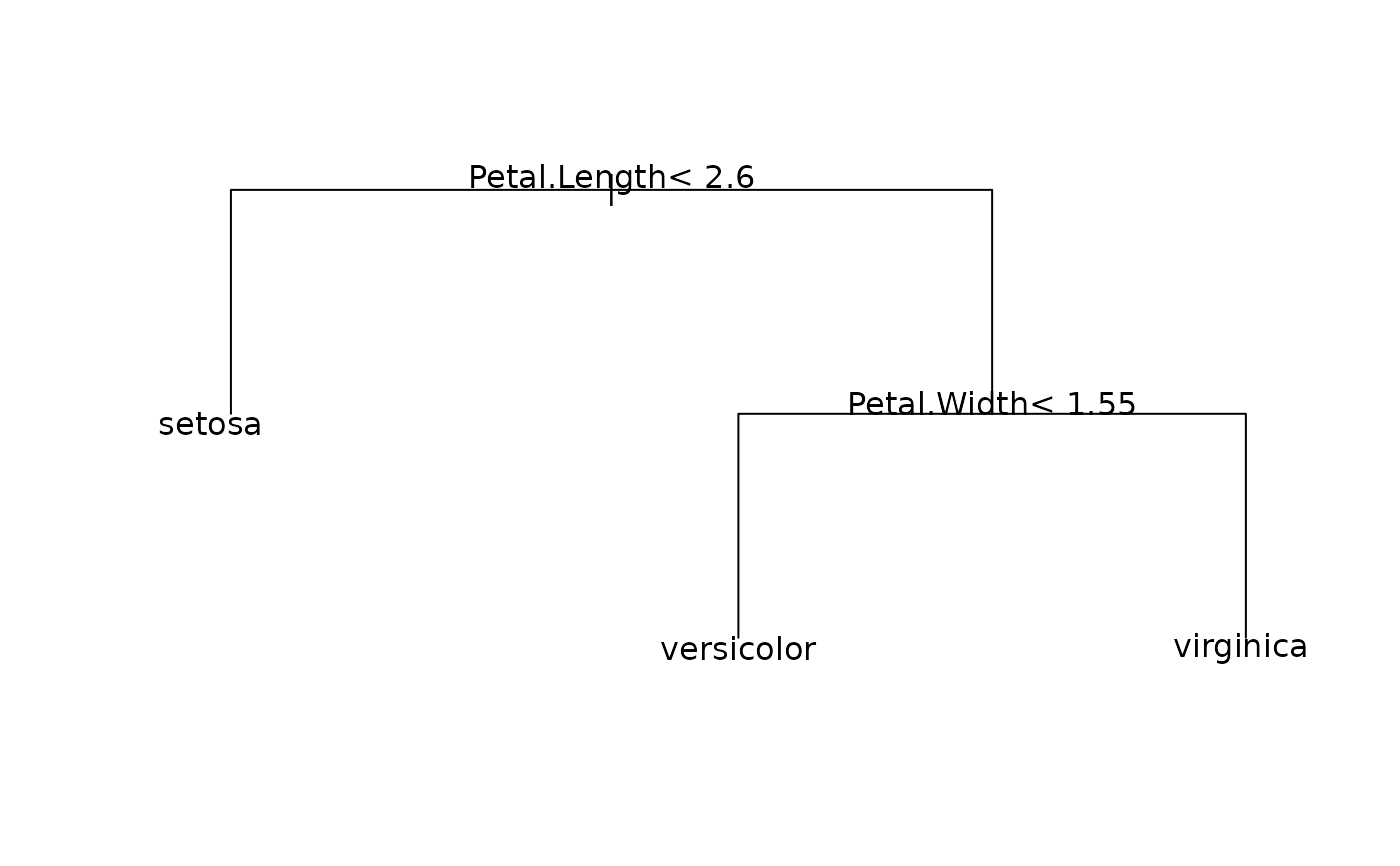 # Predictions
predict(iris_rpart) # Default type is class
#> [1] setosa setosa setosa setosa setosa setosa
#> [7] setosa setosa setosa setosa setosa setosa
#> [13] setosa setosa setosa setosa setosa setosa
#> [19] setosa setosa setosa setosa setosa setosa
#> [25] setosa setosa setosa setosa setosa setosa
#> [31] setosa setosa setosa versicolor versicolor versicolor
#> [37] versicolor versicolor versicolor virginica versicolor versicolor
#> [43] versicolor versicolor versicolor versicolor versicolor versicolor
#> [49] versicolor versicolor versicolor versicolor versicolor virginica
#> [55] versicolor versicolor versicolor versicolor versicolor versicolor
#> [61] virginica versicolor versicolor versicolor versicolor versicolor
#> [67] virginica virginica virginica virginica virginica virginica
#> [73] virginica virginica virginica virginica virginica virginica
#> [79] virginica virginica virginica virginica virginica virginica
#> [85] virginica versicolor virginica virginica virginica virginica
#> [91] virginica virginica virginica virginica virginica virginica
#> [97] virginica virginica virginica
#> Levels: setosa versicolor virginica
predict(iris_rpart, type = "membership")
#> setosa versicolor virginica
#> 2 1 0.00000000 0.00000000
#> 3 1 0.00000000 0.00000000
#> 4 1 0.00000000 0.00000000
#> 5 1 0.00000000 0.00000000
#> 6 1 0.00000000 0.00000000
#> 7 1 0.00000000 0.00000000
#> 8 1 0.00000000 0.00000000
#> 9 1 0.00000000 0.00000000
#> 10 1 0.00000000 0.00000000
#> 11 1 0.00000000 0.00000000
#> 12 1 0.00000000 0.00000000
#> 13 1 0.00000000 0.00000000
#> 14 1 0.00000000 0.00000000
#> 15 1 0.00000000 0.00000000
#> 16 1 0.00000000 0.00000000
#> 17 1 0.00000000 0.00000000
#> 18 1 0.00000000 0.00000000
#> 19 1 0.00000000 0.00000000
#> 20 1 0.00000000 0.00000000
#> 21 1 0.00000000 0.00000000
#> 22 1 0.00000000 0.00000000
#> 23 1 0.00000000 0.00000000
#> 24 1 0.00000000 0.00000000
#> 25 1 0.00000000 0.00000000
#> 26 1 0.00000000 0.00000000
#> 27 1 0.00000000 0.00000000
#> 28 1 0.00000000 0.00000000
#> 29 1 0.00000000 0.00000000
#> 30 1 0.00000000 0.00000000
#> 31 1 0.00000000 0.00000000
#> 32 1 0.00000000 0.00000000
#> 33 1 0.00000000 0.00000000
#> 34 1 0.00000000 0.00000000
#> 51 0 0.96774194 0.03225806
#> 52 0 0.96774194 0.03225806
#> 53 0 0.96774194 0.03225806
#> 54 0 0.96774194 0.03225806
#> 55 0 0.96774194 0.03225806
#> 56 0 0.96774194 0.03225806
#> 57 0 0.08571429 0.91428571
#> 58 0 0.96774194 0.03225806
#> 59 0 0.96774194 0.03225806
#> 60 0 0.96774194 0.03225806
#> 61 0 0.96774194 0.03225806
#> 62 0 0.96774194 0.03225806
#> 63 0 0.96774194 0.03225806
#> 64 0 0.96774194 0.03225806
#> 65 0 0.96774194 0.03225806
#> 66 0 0.96774194 0.03225806
#> 67 0 0.96774194 0.03225806
#> 68 0 0.96774194 0.03225806
#> 69 0 0.96774194 0.03225806
#> 70 0 0.96774194 0.03225806
#> 71 0 0.08571429 0.91428571
#> 72 0 0.96774194 0.03225806
#> 73 0 0.96774194 0.03225806
#> 74 0 0.96774194 0.03225806
#> 75 0 0.96774194 0.03225806
#> 76 0 0.96774194 0.03225806
#> 77 0 0.96774194 0.03225806
#> 78 0 0.08571429 0.91428571
#> 79 0 0.96774194 0.03225806
#> 80 0 0.96774194 0.03225806
#> 81 0 0.96774194 0.03225806
#> 82 0 0.96774194 0.03225806
#> 83 0 0.96774194 0.03225806
#> 101 0 0.08571429 0.91428571
#> 102 0 0.08571429 0.91428571
#> 103 0 0.08571429 0.91428571
#> 104 0 0.08571429 0.91428571
#> 105 0 0.08571429 0.91428571
#> 106 0 0.08571429 0.91428571
#> 107 0 0.08571429 0.91428571
#> 108 0 0.08571429 0.91428571
#> 109 0 0.08571429 0.91428571
#> 110 0 0.08571429 0.91428571
#> 111 0 0.08571429 0.91428571
#> 112 0 0.08571429 0.91428571
#> 113 0 0.08571429 0.91428571
#> 114 0 0.08571429 0.91428571
#> 115 0 0.08571429 0.91428571
#> 116 0 0.08571429 0.91428571
#> 117 0 0.08571429 0.91428571
#> 118 0 0.08571429 0.91428571
#> 119 0 0.08571429 0.91428571
#> 120 0 0.96774194 0.03225806
#> 121 0 0.08571429 0.91428571
#> 122 0 0.08571429 0.91428571
#> 123 0 0.08571429 0.91428571
#> 124 0 0.08571429 0.91428571
#> 125 0 0.08571429 0.91428571
#> 126 0 0.08571429 0.91428571
#> 127 0 0.08571429 0.91428571
#> 128 0 0.08571429 0.91428571
#> 129 0 0.08571429 0.91428571
#> 130 0 0.08571429 0.91428571
#> 131 0 0.08571429 0.91428571
#> 132 0 0.08571429 0.91428571
#> 133 0 0.08571429 0.91428571
predict(iris_rpart, type = "both")
#> $class
#> [1] setosa setosa setosa setosa setosa setosa
#> [7] setosa setosa setosa setosa setosa setosa
#> [13] setosa setosa setosa setosa setosa setosa
#> [19] setosa setosa setosa setosa setosa setosa
#> [25] setosa setosa setosa setosa setosa setosa
#> [31] setosa setosa setosa versicolor versicolor versicolor
#> [37] versicolor versicolor versicolor virginica versicolor versicolor
#> [43] versicolor versicolor versicolor versicolor versicolor versicolor
#> [49] versicolor versicolor versicolor versicolor versicolor virginica
#> [55] versicolor versicolor versicolor versicolor versicolor versicolor
#> [61] virginica versicolor versicolor versicolor versicolor versicolor
#> [67] virginica virginica virginica virginica virginica virginica
#> [73] virginica virginica virginica virginica virginica virginica
#> [79] virginica virginica virginica virginica virginica virginica
#> [85] virginica versicolor virginica virginica virginica virginica
#> [91] virginica virginica virginica virginica virginica virginica
#> [97] virginica virginica virginica
#> Levels: setosa versicolor virginica
#>
#> $membership
#> setosa versicolor virginica
#> 2 1 0.00000000 0.00000000
#> 3 1 0.00000000 0.00000000
#> 4 1 0.00000000 0.00000000
#> 5 1 0.00000000 0.00000000
#> 6 1 0.00000000 0.00000000
#> 7 1 0.00000000 0.00000000
#> 8 1 0.00000000 0.00000000
#> 9 1 0.00000000 0.00000000
#> 10 1 0.00000000 0.00000000
#> 11 1 0.00000000 0.00000000
#> 12 1 0.00000000 0.00000000
#> 13 1 0.00000000 0.00000000
#> 14 1 0.00000000 0.00000000
#> 15 1 0.00000000 0.00000000
#> 16 1 0.00000000 0.00000000
#> 17 1 0.00000000 0.00000000
#> 18 1 0.00000000 0.00000000
#> 19 1 0.00000000 0.00000000
#> 20 1 0.00000000 0.00000000
#> 21 1 0.00000000 0.00000000
#> 22 1 0.00000000 0.00000000
#> 23 1 0.00000000 0.00000000
#> 24 1 0.00000000 0.00000000
#> 25 1 0.00000000 0.00000000
#> 26 1 0.00000000 0.00000000
#> 27 1 0.00000000 0.00000000
#> 28 1 0.00000000 0.00000000
#> 29 1 0.00000000 0.00000000
#> 30 1 0.00000000 0.00000000
#> 31 1 0.00000000 0.00000000
#> 32 1 0.00000000 0.00000000
#> 33 1 0.00000000 0.00000000
#> 34 1 0.00000000 0.00000000
#> 51 0 0.96774194 0.03225806
#> 52 0 0.96774194 0.03225806
#> 53 0 0.96774194 0.03225806
#> 54 0 0.96774194 0.03225806
#> 55 0 0.96774194 0.03225806
#> 56 0 0.96774194 0.03225806
#> 57 0 0.08571429 0.91428571
#> 58 0 0.96774194 0.03225806
#> 59 0 0.96774194 0.03225806
#> 60 0 0.96774194 0.03225806
#> 61 0 0.96774194 0.03225806
#> 62 0 0.96774194 0.03225806
#> 63 0 0.96774194 0.03225806
#> 64 0 0.96774194 0.03225806
#> 65 0 0.96774194 0.03225806
#> 66 0 0.96774194 0.03225806
#> 67 0 0.96774194 0.03225806
#> 68 0 0.96774194 0.03225806
#> 69 0 0.96774194 0.03225806
#> 70 0 0.96774194 0.03225806
#> 71 0 0.08571429 0.91428571
#> 72 0 0.96774194 0.03225806
#> 73 0 0.96774194 0.03225806
#> 74 0 0.96774194 0.03225806
#> 75 0 0.96774194 0.03225806
#> 76 0 0.96774194 0.03225806
#> 77 0 0.96774194 0.03225806
#> 78 0 0.08571429 0.91428571
#> 79 0 0.96774194 0.03225806
#> 80 0 0.96774194 0.03225806
#> 81 0 0.96774194 0.03225806
#> 82 0 0.96774194 0.03225806
#> 83 0 0.96774194 0.03225806
#> 101 0 0.08571429 0.91428571
#> 102 0 0.08571429 0.91428571
#> 103 0 0.08571429 0.91428571
#> 104 0 0.08571429 0.91428571
#> 105 0 0.08571429 0.91428571
#> 106 0 0.08571429 0.91428571
#> 107 0 0.08571429 0.91428571
#> 108 0 0.08571429 0.91428571
#> 109 0 0.08571429 0.91428571
#> 110 0 0.08571429 0.91428571
#> 111 0 0.08571429 0.91428571
#> 112 0 0.08571429 0.91428571
#> 113 0 0.08571429 0.91428571
#> 114 0 0.08571429 0.91428571
#> 115 0 0.08571429 0.91428571
#> 116 0 0.08571429 0.91428571
#> 117 0 0.08571429 0.91428571
#> 118 0 0.08571429 0.91428571
#> 119 0 0.08571429 0.91428571
#> 120 0 0.96774194 0.03225806
#> 121 0 0.08571429 0.91428571
#> 122 0 0.08571429 0.91428571
#> 123 0 0.08571429 0.91428571
#> 124 0 0.08571429 0.91428571
#> 125 0 0.08571429 0.91428571
#> 126 0 0.08571429 0.91428571
#> 127 0 0.08571429 0.91428571
#> 128 0 0.08571429 0.91428571
#> 129 0 0.08571429 0.91428571
#> 130 0 0.08571429 0.91428571
#> 131 0 0.08571429 0.91428571
#> 132 0 0.08571429 0.91428571
#> 133 0 0.08571429 0.91428571
#>
# Self-consistency, do not use for assessing classifier performances!
confusion(iris_rpart)
#> 99 items classified with 95 true positives (error rate = 4%)
#> Predicted
#> Actual 01 02 03 (sum) (FNR%)
#> 01 setosa 33 0 0 33 0
#> 02 versicolor 0 30 3 33 9
#> 03 virginica 0 1 32 33 3
#> (sum) 33 31 35 99 4
# Cross-validation prediction is a good choice when there is no test set
predict(iris_rpart, method = "cv") # Idem: cvpredict(res)
#> [1] setosa setosa setosa setosa setosa setosa
#> [7] setosa setosa setosa setosa setosa setosa
#> [13] setosa setosa setosa setosa setosa setosa
#> [19] setosa setosa setosa setosa setosa setosa
#> [25] setosa setosa setosa setosa setosa setosa
#> [31] setosa setosa setosa versicolor versicolor versicolor
#> [37] versicolor versicolor versicolor virginica versicolor versicolor
#> [43] versicolor versicolor versicolor versicolor versicolor versicolor
#> [49] versicolor versicolor versicolor versicolor versicolor virginica
#> [55] versicolor versicolor versicolor versicolor versicolor versicolor
#> [61] virginica versicolor versicolor versicolor versicolor versicolor
#> [67] virginica virginica virginica virginica virginica virginica
#> [73] versicolor virginica virginica virginica virginica virginica
#> [79] virginica virginica virginica virginica virginica virginica
#> [85] virginica versicolor virginica virginica virginica virginica
#> [91] virginica virginica virginica virginica virginica versicolor
#> [97] virginica virginica virginica
#> attr(,"method")
#>
#> Call:
#> cvpredict.mlearning(object = object, type = type)
#>
#> 10-fold cross-validation estimator of misclassification error
#>
#> Misclassification error: 0.0606
#>
#> Levels: setosa versicolor virginica
confusion(iris_rpart, method = "cv")
#> 99 items classified with 90 true positives (error rate = 9.1%)
#> Predicted
#> Actual 01 02 03 (sum) (FNR%)
#> 01 setosa 33 0 0 33 0
#> 02 versicolor 0 28 5 33 15
#> 03 virginica 0 4 29 33 12
#> (sum) 33 32 34 99 9
# Evaluation of performances using a separate test set
confusion(predict(iris_rpart, newdata = iris_test), iris_test$Species)
#> 50 items classified with 45 true positives (error rate = 10%)
#> Predicted
#> Actual 01 02 03 04 (sum) (FNR%)
#> 01 versicolor 14 2 0 1 17 18
#> 02 virginica 2 15 0 0 17 12
#> 03 setosa 0 0 16 0 16 0
#> 04 NA 0 0 0 0 0
#> (sum) 16 17 16 1 50 10
# Predictions
predict(iris_rpart) # Default type is class
#> [1] setosa setosa setosa setosa setosa setosa
#> [7] setosa setosa setosa setosa setosa setosa
#> [13] setosa setosa setosa setosa setosa setosa
#> [19] setosa setosa setosa setosa setosa setosa
#> [25] setosa setosa setosa setosa setosa setosa
#> [31] setosa setosa setosa versicolor versicolor versicolor
#> [37] versicolor versicolor versicolor virginica versicolor versicolor
#> [43] versicolor versicolor versicolor versicolor versicolor versicolor
#> [49] versicolor versicolor versicolor versicolor versicolor virginica
#> [55] versicolor versicolor versicolor versicolor versicolor versicolor
#> [61] virginica versicolor versicolor versicolor versicolor versicolor
#> [67] virginica virginica virginica virginica virginica virginica
#> [73] virginica virginica virginica virginica virginica virginica
#> [79] virginica virginica virginica virginica virginica virginica
#> [85] virginica versicolor virginica virginica virginica virginica
#> [91] virginica virginica virginica virginica virginica virginica
#> [97] virginica virginica virginica
#> Levels: setosa versicolor virginica
predict(iris_rpart, type = "membership")
#> setosa versicolor virginica
#> 2 1 0.00000000 0.00000000
#> 3 1 0.00000000 0.00000000
#> 4 1 0.00000000 0.00000000
#> 5 1 0.00000000 0.00000000
#> 6 1 0.00000000 0.00000000
#> 7 1 0.00000000 0.00000000
#> 8 1 0.00000000 0.00000000
#> 9 1 0.00000000 0.00000000
#> 10 1 0.00000000 0.00000000
#> 11 1 0.00000000 0.00000000
#> 12 1 0.00000000 0.00000000
#> 13 1 0.00000000 0.00000000
#> 14 1 0.00000000 0.00000000
#> 15 1 0.00000000 0.00000000
#> 16 1 0.00000000 0.00000000
#> 17 1 0.00000000 0.00000000
#> 18 1 0.00000000 0.00000000
#> 19 1 0.00000000 0.00000000
#> 20 1 0.00000000 0.00000000
#> 21 1 0.00000000 0.00000000
#> 22 1 0.00000000 0.00000000
#> 23 1 0.00000000 0.00000000
#> 24 1 0.00000000 0.00000000
#> 25 1 0.00000000 0.00000000
#> 26 1 0.00000000 0.00000000
#> 27 1 0.00000000 0.00000000
#> 28 1 0.00000000 0.00000000
#> 29 1 0.00000000 0.00000000
#> 30 1 0.00000000 0.00000000
#> 31 1 0.00000000 0.00000000
#> 32 1 0.00000000 0.00000000
#> 33 1 0.00000000 0.00000000
#> 34 1 0.00000000 0.00000000
#> 51 0 0.96774194 0.03225806
#> 52 0 0.96774194 0.03225806
#> 53 0 0.96774194 0.03225806
#> 54 0 0.96774194 0.03225806
#> 55 0 0.96774194 0.03225806
#> 56 0 0.96774194 0.03225806
#> 57 0 0.08571429 0.91428571
#> 58 0 0.96774194 0.03225806
#> 59 0 0.96774194 0.03225806
#> 60 0 0.96774194 0.03225806
#> 61 0 0.96774194 0.03225806
#> 62 0 0.96774194 0.03225806
#> 63 0 0.96774194 0.03225806
#> 64 0 0.96774194 0.03225806
#> 65 0 0.96774194 0.03225806
#> 66 0 0.96774194 0.03225806
#> 67 0 0.96774194 0.03225806
#> 68 0 0.96774194 0.03225806
#> 69 0 0.96774194 0.03225806
#> 70 0 0.96774194 0.03225806
#> 71 0 0.08571429 0.91428571
#> 72 0 0.96774194 0.03225806
#> 73 0 0.96774194 0.03225806
#> 74 0 0.96774194 0.03225806
#> 75 0 0.96774194 0.03225806
#> 76 0 0.96774194 0.03225806
#> 77 0 0.96774194 0.03225806
#> 78 0 0.08571429 0.91428571
#> 79 0 0.96774194 0.03225806
#> 80 0 0.96774194 0.03225806
#> 81 0 0.96774194 0.03225806
#> 82 0 0.96774194 0.03225806
#> 83 0 0.96774194 0.03225806
#> 101 0 0.08571429 0.91428571
#> 102 0 0.08571429 0.91428571
#> 103 0 0.08571429 0.91428571
#> 104 0 0.08571429 0.91428571
#> 105 0 0.08571429 0.91428571
#> 106 0 0.08571429 0.91428571
#> 107 0 0.08571429 0.91428571
#> 108 0 0.08571429 0.91428571
#> 109 0 0.08571429 0.91428571
#> 110 0 0.08571429 0.91428571
#> 111 0 0.08571429 0.91428571
#> 112 0 0.08571429 0.91428571
#> 113 0 0.08571429 0.91428571
#> 114 0 0.08571429 0.91428571
#> 115 0 0.08571429 0.91428571
#> 116 0 0.08571429 0.91428571
#> 117 0 0.08571429 0.91428571
#> 118 0 0.08571429 0.91428571
#> 119 0 0.08571429 0.91428571
#> 120 0 0.96774194 0.03225806
#> 121 0 0.08571429 0.91428571
#> 122 0 0.08571429 0.91428571
#> 123 0 0.08571429 0.91428571
#> 124 0 0.08571429 0.91428571
#> 125 0 0.08571429 0.91428571
#> 126 0 0.08571429 0.91428571
#> 127 0 0.08571429 0.91428571
#> 128 0 0.08571429 0.91428571
#> 129 0 0.08571429 0.91428571
#> 130 0 0.08571429 0.91428571
#> 131 0 0.08571429 0.91428571
#> 132 0 0.08571429 0.91428571
#> 133 0 0.08571429 0.91428571
predict(iris_rpart, type = "both")
#> $class
#> [1] setosa setosa setosa setosa setosa setosa
#> [7] setosa setosa setosa setosa setosa setosa
#> [13] setosa setosa setosa setosa setosa setosa
#> [19] setosa setosa setosa setosa setosa setosa
#> [25] setosa setosa setosa setosa setosa setosa
#> [31] setosa setosa setosa versicolor versicolor versicolor
#> [37] versicolor versicolor versicolor virginica versicolor versicolor
#> [43] versicolor versicolor versicolor versicolor versicolor versicolor
#> [49] versicolor versicolor versicolor versicolor versicolor virginica
#> [55] versicolor versicolor versicolor versicolor versicolor versicolor
#> [61] virginica versicolor versicolor versicolor versicolor versicolor
#> [67] virginica virginica virginica virginica virginica virginica
#> [73] virginica virginica virginica virginica virginica virginica
#> [79] virginica virginica virginica virginica virginica virginica
#> [85] virginica versicolor virginica virginica virginica virginica
#> [91] virginica virginica virginica virginica virginica virginica
#> [97] virginica virginica virginica
#> Levels: setosa versicolor virginica
#>
#> $membership
#> setosa versicolor virginica
#> 2 1 0.00000000 0.00000000
#> 3 1 0.00000000 0.00000000
#> 4 1 0.00000000 0.00000000
#> 5 1 0.00000000 0.00000000
#> 6 1 0.00000000 0.00000000
#> 7 1 0.00000000 0.00000000
#> 8 1 0.00000000 0.00000000
#> 9 1 0.00000000 0.00000000
#> 10 1 0.00000000 0.00000000
#> 11 1 0.00000000 0.00000000
#> 12 1 0.00000000 0.00000000
#> 13 1 0.00000000 0.00000000
#> 14 1 0.00000000 0.00000000
#> 15 1 0.00000000 0.00000000
#> 16 1 0.00000000 0.00000000
#> 17 1 0.00000000 0.00000000
#> 18 1 0.00000000 0.00000000
#> 19 1 0.00000000 0.00000000
#> 20 1 0.00000000 0.00000000
#> 21 1 0.00000000 0.00000000
#> 22 1 0.00000000 0.00000000
#> 23 1 0.00000000 0.00000000
#> 24 1 0.00000000 0.00000000
#> 25 1 0.00000000 0.00000000
#> 26 1 0.00000000 0.00000000
#> 27 1 0.00000000 0.00000000
#> 28 1 0.00000000 0.00000000
#> 29 1 0.00000000 0.00000000
#> 30 1 0.00000000 0.00000000
#> 31 1 0.00000000 0.00000000
#> 32 1 0.00000000 0.00000000
#> 33 1 0.00000000 0.00000000
#> 34 1 0.00000000 0.00000000
#> 51 0 0.96774194 0.03225806
#> 52 0 0.96774194 0.03225806
#> 53 0 0.96774194 0.03225806
#> 54 0 0.96774194 0.03225806
#> 55 0 0.96774194 0.03225806
#> 56 0 0.96774194 0.03225806
#> 57 0 0.08571429 0.91428571
#> 58 0 0.96774194 0.03225806
#> 59 0 0.96774194 0.03225806
#> 60 0 0.96774194 0.03225806
#> 61 0 0.96774194 0.03225806
#> 62 0 0.96774194 0.03225806
#> 63 0 0.96774194 0.03225806
#> 64 0 0.96774194 0.03225806
#> 65 0 0.96774194 0.03225806
#> 66 0 0.96774194 0.03225806
#> 67 0 0.96774194 0.03225806
#> 68 0 0.96774194 0.03225806
#> 69 0 0.96774194 0.03225806
#> 70 0 0.96774194 0.03225806
#> 71 0 0.08571429 0.91428571
#> 72 0 0.96774194 0.03225806
#> 73 0 0.96774194 0.03225806
#> 74 0 0.96774194 0.03225806
#> 75 0 0.96774194 0.03225806
#> 76 0 0.96774194 0.03225806
#> 77 0 0.96774194 0.03225806
#> 78 0 0.08571429 0.91428571
#> 79 0 0.96774194 0.03225806
#> 80 0 0.96774194 0.03225806
#> 81 0 0.96774194 0.03225806
#> 82 0 0.96774194 0.03225806
#> 83 0 0.96774194 0.03225806
#> 101 0 0.08571429 0.91428571
#> 102 0 0.08571429 0.91428571
#> 103 0 0.08571429 0.91428571
#> 104 0 0.08571429 0.91428571
#> 105 0 0.08571429 0.91428571
#> 106 0 0.08571429 0.91428571
#> 107 0 0.08571429 0.91428571
#> 108 0 0.08571429 0.91428571
#> 109 0 0.08571429 0.91428571
#> 110 0 0.08571429 0.91428571
#> 111 0 0.08571429 0.91428571
#> 112 0 0.08571429 0.91428571
#> 113 0 0.08571429 0.91428571
#> 114 0 0.08571429 0.91428571
#> 115 0 0.08571429 0.91428571
#> 116 0 0.08571429 0.91428571
#> 117 0 0.08571429 0.91428571
#> 118 0 0.08571429 0.91428571
#> 119 0 0.08571429 0.91428571
#> 120 0 0.96774194 0.03225806
#> 121 0 0.08571429 0.91428571
#> 122 0 0.08571429 0.91428571
#> 123 0 0.08571429 0.91428571
#> 124 0 0.08571429 0.91428571
#> 125 0 0.08571429 0.91428571
#> 126 0 0.08571429 0.91428571
#> 127 0 0.08571429 0.91428571
#> 128 0 0.08571429 0.91428571
#> 129 0 0.08571429 0.91428571
#> 130 0 0.08571429 0.91428571
#> 131 0 0.08571429 0.91428571
#> 132 0 0.08571429 0.91428571
#> 133 0 0.08571429 0.91428571
#>
# Self-consistency, do not use for assessing classifier performances!
confusion(iris_rpart)
#> 99 items classified with 95 true positives (error rate = 4%)
#> Predicted
#> Actual 01 02 03 (sum) (FNR%)
#> 01 setosa 33 0 0 33 0
#> 02 versicolor 0 30 3 33 9
#> 03 virginica 0 1 32 33 3
#> (sum) 33 31 35 99 4
# Cross-validation prediction is a good choice when there is no test set
predict(iris_rpart, method = "cv") # Idem: cvpredict(res)
#> [1] setosa setosa setosa setosa setosa setosa
#> [7] setosa setosa setosa setosa setosa setosa
#> [13] setosa setosa setosa setosa setosa setosa
#> [19] setosa setosa setosa setosa setosa setosa
#> [25] setosa setosa setosa setosa setosa setosa
#> [31] setosa setosa setosa versicolor versicolor versicolor
#> [37] versicolor versicolor versicolor virginica versicolor versicolor
#> [43] versicolor versicolor versicolor versicolor versicolor versicolor
#> [49] versicolor versicolor versicolor versicolor versicolor virginica
#> [55] versicolor versicolor versicolor versicolor versicolor versicolor
#> [61] virginica versicolor versicolor versicolor versicolor versicolor
#> [67] virginica virginica virginica virginica virginica virginica
#> [73] versicolor virginica virginica virginica virginica virginica
#> [79] virginica virginica virginica virginica virginica virginica
#> [85] virginica versicolor virginica virginica virginica virginica
#> [91] virginica virginica virginica virginica virginica versicolor
#> [97] virginica virginica virginica
#> attr(,"method")
#>
#> Call:
#> cvpredict.mlearning(object = object, type = type)
#>
#> 10-fold cross-validation estimator of misclassification error
#>
#> Misclassification error: 0.0606
#>
#> Levels: setosa versicolor virginica
confusion(iris_rpart, method = "cv")
#> 99 items classified with 90 true positives (error rate = 9.1%)
#> Predicted
#> Actual 01 02 03 (sum) (FNR%)
#> 01 setosa 33 0 0 33 0
#> 02 versicolor 0 28 5 33 15
#> 03 virginica 0 4 29 33 12
#> (sum) 33 32 34 99 9
# Evaluation of performances using a separate test set
confusion(predict(iris_rpart, newdata = iris_test), iris_test$Species)
#> 50 items classified with 45 true positives (error rate = 10%)
#> Predicted
#> Actual 01 02 03 04 (sum) (FNR%)
#> 01 versicolor 14 2 0 1 17 18
#> 02 virginica 2 15 0 0 17 12
#> 03 setosa 0 0 16 0 16 0
#> 04 NA 0 0 0 0 0
#> (sum) 16 17 16 1 50 10
